-Search query
-Search result
Showing 1 - 50 of 135 items for (author: huang & sj)

EMDB-17360: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP
Method: single particle / : Banerjee I, Bimai O, Sjoberg BM, Logan DT

EMDB-17361: 
Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP
Method: single particle / : Banerjee I, Bimai O, Sjoberg BM, Logan DT

EMDB-17373: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/dTTP/GTP-bound state
Method: single particle / : Bimai O, Banerjee I, Sjoberg BM, Logan DT

EMDB-17385: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state
Method: single particle / : Bimai O, Banerjee I, Sjoberg BM, Logan DT

EMDB-17357: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/CTP-bound state
Method: single particle / : Banerjee I, Bimai O, Sjoberg BM, Logan DT

EMDB-17358: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dATP-bound state
Method: single particle / : Banerjee I, Bimai O, Sjoberg BM, Logan DT

EMDB-17359: 
Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric, dATP-bound state
Method: single particle / : Banerjee I, Bimai O, Sjoberg BM, Logan DT
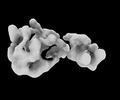
EMDB-35767: 
Structure of the Mex67-Mtr2-3 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34638: 
Structure of the Mex67-Mtr2-1 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF
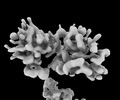
EMDB-34640: 
Structure of Mex67-Mtr2-2 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF
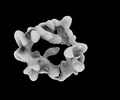
EMDB-34641: 
Structure of Crm1-RanGTP complex
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34725: 
NPC-trapped pre-60S particle
Method: single particle / : Li ZQ, Chen SJ, Sui SF
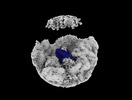
EMDB-35812: 
Bud20 interacts with CFNC
Method: single particle / : Li ZQ, Chen SJ

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
Method: single particle / : Shek J, Callaway H, Li H, Yu X, Saphire EO
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28096: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28097: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
Method: single particle / : Yu X, Callaway H, Li H, Shek J, Saphire EO
EMDB-28098: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-294
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28099: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-295
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28100: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-299
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28102: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-334
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO
EMDB-28103: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-360
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28104: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-361
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28105: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-362
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28106: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-368
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28168: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-292
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28169: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-333
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO
EMDB-28170: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-355
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28171: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-371
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-16022: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordberg A, Goedert M, Scheres SHW

EMDB-16023: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

EMDB-16027: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

PDB-8bfz: 
Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordberg A, Goedert M, Scheres SHW

PDB-8bg0: 
Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

PDB-8bg9: 
Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta
Method: helical / : Yang Y, Zhang WJ, Murzin AG, Schweighauser M, Huang M, Lovestam SKA, Peak-Chew SY, Macdonald J, Lavenir I, Ghetti B, Graff C, Kumar A, Nordber A, Goedert M, Scheres SHW

EMDB-16434: 
Type Ib beta-amyloid 42 Filaments from Human Brain
Method: helical / : Yang Y, Arseni D, Zhang W, Huang M, Lovestam SKA, Schweighauser M, Kotecha A, Murzin AG, Peak-Chew SY, Macdonald J, Lavenir I, Garringer HJ, Gelpi E, Newell KL, Kovacs GG, Vidal R, Ghetti B, Falcon B, Scheres HW, Goedert M

EMDB-33233: 
Cryo-EM structure of EDS1 and SAG101 with ATP-APDR
Method: single particle / : Huang SJ, Jia AL, Han ZF, Chai JJ

EMDB-33144: 
Cryo-EM structure of EDS1 and PAD4
Method: single particle / : Huang SJ, Jia AL, Sun Y, Han ZF, Chai JJ

EMDB-32653: 
Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex
Method: single particle / : Li ZQ, Chen SJB, Zhao L, Sui SF

EMDB-32658: 
Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex
Method: single particle / : Li ZQ, Chen SJB, Zhao L, Sui SF

EMDB-32662: 
Cryo-EM map of the inner ring dimer of the Saccharomyces cerevisiae nuclear pore complex
Method: single particle / : Li ZQ, Chen SJB, Zhao L, Sui SF
EMDB-32663: 
Cryo-EM map of the intact inner ring of the Saccharomyces cerevisiae nuclear pore complex
Method: single particle / : Li ZQ, Chen SJB, Zhao L, Sui SF

EMDB-32664: 
Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex
Method: single particle / : Li ZQ, Chen SJB, Zhao L, Sui SF

EMDB-32684: 
SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab
Method: single particle / : Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, Wu K, Hu W, Huang Z, Liu J, Wang Z, Jia F, Xia K, Liu P, Song B, Zheng J, Jiang H, Cheng X, Jiang Y, Deng SJ, Xu HE

EMDB-32736: 
The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S
Method: single particle / : Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, Wu K, Hu W, Huang Z, Liu J, Wang Z, Jia F, Xia K, Liu P, Song B, Zheng J, Jiang H, Cheng X, Jiang Y, Deng SJ, Xu HE

EMDB-32683: 
SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab
Method: single particle / : Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, Wu K, Hu W, Huang Z, Liu J, Wang Z, Jia F, Xia K, Liu P, Song B, Zheng J, Jiang H, Cheng X, Jiang Y, Deng SJ, Xu HE
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
